ISSN-Online: 2313-7177; Frequency: Annual, Instant published; The first Issue: March, 2015; Full Open Access; Publishing fee
SCOPE: JBMBR publishes original research, reviews and other material related to Bioenergetics, Cell Biology, Chromosomes, Developmental biology, DNA, Enzymology, Extracellular Matrices, Gene Regulation, Genomics, Glycobiology, Immunology, Lipids, Membrane Biology, Membrane Biology, Metabolism, Molecular Bases of Disease, Molecular Biophysics, Neurobiology, Plant Biology, Protein Structure and Folding, Protein Synthesis and Degradation, Proteomics, Signal Transduction, DNA replication, repair and recombination, gene expression, epigenetics and chromatin structure and function, RNA processing, functions of non coding RNAs, transcription, Structure, chemistry, processing and function of biologically important macromolecules and complexes, Biomolecular interactions, systems biology, Computational biology, Translation, protein folding, processing and degradation, Sorting, spatiotemporal organization, trafficking, signal transduction and intracellular signaling, Membrane processes, cell surface proteins and cell-cell interactions, Molecular basis of disease, Methodological advances, both experimental and theoretical, including databases.
MISSION: Rapid exchange of scientific information between clinicians and scientists worldwide, seeks to publish papers reporting original clinical and scientific research which are of a high standard and which contribute to the advancement of knowledge in the field of biological chemistry and molecular biology.
EDITOR-IN-CHIEF
Ira-Ida Skvortsova, Head of the Laboratory for Experimental and Translational Resaerch on Radiation Oncology (EXTRO-Lab) in the Department of Therapeutic Radiology and Oncology at Innsbruck Medical University, Innsbruck, Austria.
MoreCOVER IMAGE
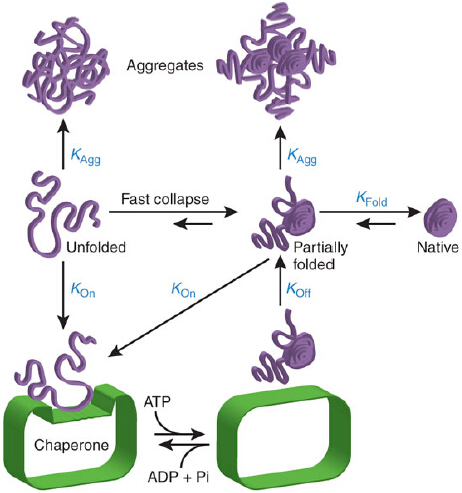
Figure 1: Chaperone mechanism in promoting folding through kinetic of partitioning.Reprinted with permission from Hartl FU,Hayer-Hartl M 2009. Copyright 2009 Nature Structural and Molecular Biology.
MoreFeatured
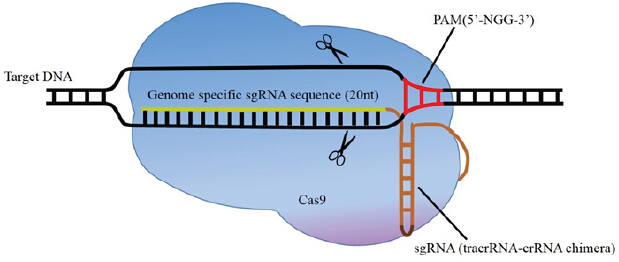 Figure 1: Illustration of CRISPR/Cas9-mediated genome editing. The dual tracrRNA:crRNA is engineered as a single guide RNA (sgRNA), which is a chimeric RNA containing all essential crRNA and tracrRNA components. sgRNA guides Cas9 to a target site when detects complementarity between a “protospacer” sequence in the target DNA and the remaining spacer sequence in the crRNA. After binding to the target site, the DNA single-strand matching crRNA and opposite strand are cleaved by the HNH nuclease domain and RuvC-like nuclease domain of Cas9 respectively, generating a double-strand break (DSB). ABSTRACT: Genetically modified cells and animals represent a crucial tool for understanding gene function in development and diseases. The recently developed simple-to-design, easy-to-use and multiplexing CRISPR/Cas9 system is an efficient gene-targeting technology that has already stimulated innovative applications in biology and enabled researchers to make changes in the sequence or expression of any gene in virtually any cell type or organism of interest. When combined with large libraries of sgRNAs, CRISPR/Cas9 enables facile comprehensive forward genetic screens both in vitro and in vivo. Although challenges still remain regarding the off-target mutations, delivery methods as well as the frequency of homology-directed repair, the rapid advance in CRISPR/Cas9 technology opens the door for gene function revealing and genome and epigenome editing. It can be optimistically anticipated that, in the future, CRISPR/Cas9 technology may revolutionize gene therapy research and become a convenient and versatile tool to treat a wide variety of human diseases.More
Figure 1: Illustration of CRISPR/Cas9-mediated genome editing. The dual tracrRNA:crRNA is engineered as a single guide RNA (sgRNA), which is a chimeric RNA containing all essential crRNA and tracrRNA components. sgRNA guides Cas9 to a target site when detects complementarity between a “protospacer” sequence in the target DNA and the remaining spacer sequence in the crRNA. After binding to the target site, the DNA single-strand matching crRNA and opposite strand are cleaved by the HNH nuclease domain and RuvC-like nuclease domain of Cas9 respectively, generating a double-strand break (DSB). ABSTRACT: Genetically modified cells and animals represent a crucial tool for understanding gene function in development and diseases. The recently developed simple-to-design, easy-to-use and multiplexing CRISPR/Cas9 system is an efficient gene-targeting technology that has already stimulated innovative applications in biology and enabled researchers to make changes in the sequence or expression of any gene in virtually any cell type or organism of interest. When combined with large libraries of sgRNAs, CRISPR/Cas9 enables facile comprehensive forward genetic screens both in vitro and in vivo. Although challenges still remain regarding the off-target mutations, delivery methods as well as the frequency of homology-directed repair, the rapid advance in CRISPR/Cas9 technology opens the door for gene function revealing and genome and epigenome editing. It can be optimistically anticipated that, in the future, CRISPR/Cas9 technology may revolutionize gene therapy research and become a convenient and versatile tool to treat a wide variety of human diseases.More
Current
EXCELLENT ARTICLES
Table of Contents
EDITORIAL
| Bridged Nucleic Acids (BNAs) as Molecular Tools |
PDF
HTML
|
|
Sung-Kun Kim, Klaus D Linse, Parker Retes, Patrick Castro, Miguel Castro |
67-71 |
REVIEW
ORIGINAL ARTICLE
| Autophagy and Cellular Senescence in Lung Diseases |
PDF
HTML
|
|
Kazuyoshi Kuwano, Jun Araya, Hiromichi Hara, Shunsuke Minagawa, Naoki Takasaka, Saburo Ito, Katsutoshi Nakayama |
54-66 |
Articles
ISSN: 2313-7177
 Ira-Ida Skvortsova, Head of the Laboratory for Experimental and Translational Resaerch on Radiation Oncology (EXTRO-Lab) in the Department of Therapeutic Radiology and Oncology at Innsbruck Medical University, Innsbruck, Austria. More
Ira-Ida Skvortsova, Head of the Laboratory for Experimental and Translational Resaerch on Radiation Oncology (EXTRO-Lab) in the Department of Therapeutic Radiology and Oncology at Innsbruck Medical University, Innsbruck, Austria. More Figure 1: Chaperone mechanism in promoting folding through kinetic of partitioning.Reprinted with permission from Hartl FU,Hayer-Hartl M 2009. Copyright 2009 Nature Structural and Molecular Biology. More
Figure 1: Chaperone mechanism in promoting folding through kinetic of partitioning.Reprinted with permission from Hartl FU,Hayer-Hartl M 2009. Copyright 2009 Nature Structural and Molecular Biology. More Figure 1: Illustration of CRISPR/Cas9-mediated genome editing. The dual tracrRNA:crRNA is engineered as a single guide RNA (sgRNA), which is a chimeric RNA containing all essential crRNA and tracrRNA components. sgRNA guides Cas9 to a target site when detects complementarity between a “protospacer” sequence in the target DNA and the remaining spacer sequence in the crRNA. After binding to the target site, the DNA single-strand matching crRNA and opposite strand are cleaved by the HNH nuclease domain and RuvC-like nuclease domain of Cas9 respectively, generating a double-strand break (DSB). ABSTRACT: Genetically modified cells and animals represent a crucial tool for understanding gene function in development and diseases. The recently developed simple-to-design, easy-to-use and multiplexing CRISPR/Cas9 system is an efficient gene-targeting technology that has already stimulated innovative applications in biology and enabled researchers to make changes in the sequence or expression of any gene in virtually any cell type or organism of interest. When combined with large libraries of sgRNAs, CRISPR/Cas9 enables facile comprehensive forward genetic screens both in vitro and in vivo. Although challenges still remain regarding the off-target mutations, delivery methods as well as the frequency of homology-directed repair, the rapid advance in CRISPR/Cas9 technology opens the door for gene function revealing and genome and epigenome editing. It can be optimistically anticipated that, in the future, CRISPR/Cas9 technology may revolutionize gene therapy research and become a convenient and versatile tool to treat a wide variety of human diseases.More
Figure 1: Illustration of CRISPR/Cas9-mediated genome editing. The dual tracrRNA:crRNA is engineered as a single guide RNA (sgRNA), which is a chimeric RNA containing all essential crRNA and tracrRNA components. sgRNA guides Cas9 to a target site when detects complementarity between a “protospacer” sequence in the target DNA and the remaining spacer sequence in the crRNA. After binding to the target site, the DNA single-strand matching crRNA and opposite strand are cleaved by the HNH nuclease domain and RuvC-like nuclease domain of Cas9 respectively, generating a double-strand break (DSB). ABSTRACT: Genetically modified cells and animals represent a crucial tool for understanding gene function in development and diseases. The recently developed simple-to-design, easy-to-use and multiplexing CRISPR/Cas9 system is an efficient gene-targeting technology that has already stimulated innovative applications in biology and enabled researchers to make changes in the sequence or expression of any gene in virtually any cell type or organism of interest. When combined with large libraries of sgRNAs, CRISPR/Cas9 enables facile comprehensive forward genetic screens both in vitro and in vivo. Although challenges still remain regarding the off-target mutations, delivery methods as well as the frequency of homology-directed repair, the rapid advance in CRISPR/Cas9 technology opens the door for gene function revealing and genome and epigenome editing. It can be optimistically anticipated that, in the future, CRISPR/Cas9 technology may revolutionize gene therapy research and become a convenient and versatile tool to treat a wide variety of human diseases.More 


